
[ad_1]
In a current research posted to the bioRxiv* pre-print server, researchers characterised the extreme acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VOC) Omicron’s new sub-lineages, BA.4, BA.5, and BA.3. They reported the antigenic characterization of those sublineages in comparison with the beforehand recognized Omicron sublineages, BA.1 and BA.2.
 Research: Additional antibody escape by Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Picture Credit score: NIAID
Research: Additional antibody escape by Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Picture Credit score: NIAID
Background
Omicron sublineages, BA.4 and BA.5, have been reported from Gauteng in South Africa in April 2022. It is doable that they’re fueling a brand new wave of infections in South Africa, which may ultimately erupt worldwide. Each BA.4/5 seem to have advanced from BA.2 and have comparable SARS-CoV-2 spike (S) glycoprotein sequences. Moreover, they include mutations within the S receptor-binding area (RBD), viz., the R493Q mutation (additionally discovered within the SARS-CoV-2 Wuhan-Hu 1 pressure), in addition to L452R and F486V substitutions.
The substitutions within the BA.4/5 RBDs, L452R, and F486V are of most concern due to their potential to confer immune invasion. Each the mutations are near the angiotensin-converting enzyme 2 (ACE2) receptor floor, therefore can modulate RBD-ACE2 affinity and the neutralizing capability of pure or vaccine acquired immunity. Whereas the reversion mutation Q493, which additionally lies inside the ACE2, probably reduces the escape from responses to earlier SARS-CoV-2 strains.
In regards to the research
Within the current research, researchers reported antigenic traits of the Omicron sub-lineages BA.4 and BA.5 (BA.4/5). To this finish, the staff constructed pseudotyped lentiviruses (pLVs) expressing the S gene of the Omicron sub-lineages BA.1, BA.1.1, BA.2, BA.3, and BA.4/5. They used Wuhan-Hu 1 associated pressure, Victoria, because the management.
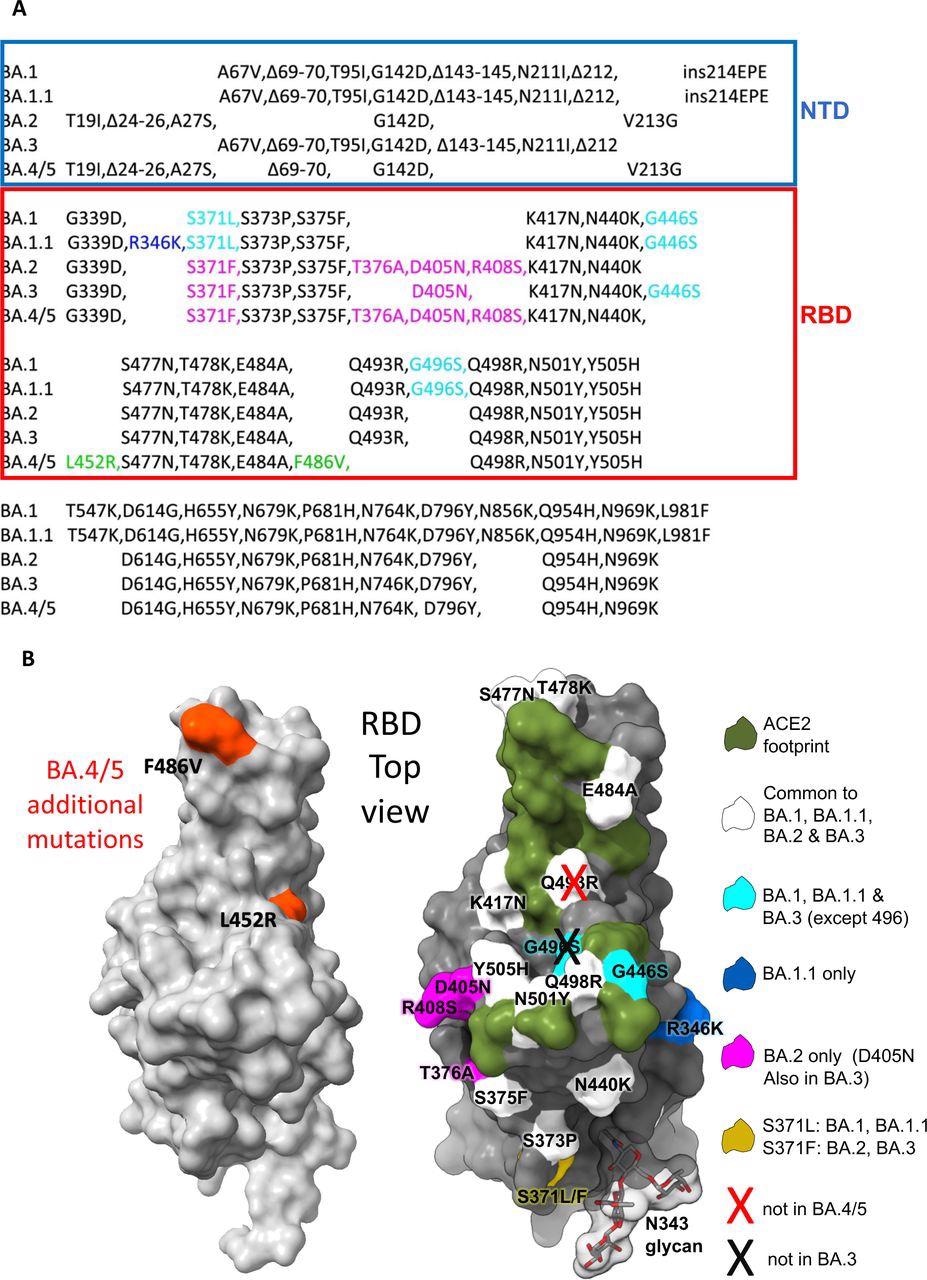
The Omicron sub-lineage in comparison with BA.4/5. (A) Comparability of S protein mutations of Omicron BA.1, BA.1.1, BA.2, BA.3 and BA.4/5 with NTD and RBD boundaries indicated. (B) Place of RBD mutations (gray floor with the ACE2 footprint in darkish inexperienced). Mutations widespread to all Omicron lineages are proven in white (Q493R which is reverted in BA.4/5 is proven with a cross), these widespread to BA.1 and BA.1.1 in cyan, these distinctive to BA.1.1 in blue and people distinctive to BA.2 in magenta. Residue 371 (yellow) is mutated in all Omicron viruses however differs between BA.1 and BA.2. The N343 glycan is proven as sticks with a clear floor.
Subsequent, the staff collected serum samples of coronavirus illness 2019 (COVID-19) vaccinated people to carry out neutralization assays. Particularly, they obtained 41 and 20 serum samples 28 days following the third dose of the AZD1222 and BNT162b2 vaccine, respectively.
Additional, the staff collected serum samples from people who had suffered breakthrough Omicron infections at two-time factors. Accordingly, they collected samples after 14 days from symptom onset and 21 days from symptom onset.
Moreover, the researchers used floor plasmon resonance (SPR) to check the binding of BA.4/5 and BA.2 RBDs to a panel of therapeutic antibodies. Lastly, the researchers used the neutralization knowledge to put BA.3 and BA.4/5 on an antigenic map.
Research findings
Nutalai et al. have demonstrated that Omicron BA.1 an infection following vaccination results in induction of broadly neutralizing antibodies, with excessive titers towards all of the VOCs. In distinction, on the early time level, BA.4/5 titers have been lowered 1.9-fold and 1.5-fold in comparison with BA.1 and BA.2, respectively. On the later time level, BA.4/5 titers have been lowered 3.4-fold and two-fold in comparison with BA.1 and BA.2, respectively. The observations point out that BA.4/5 escaped each the vaccine-induced and BA.1 infection-induced antibodies greater than BA.1 and BA.2 sub-lineages.
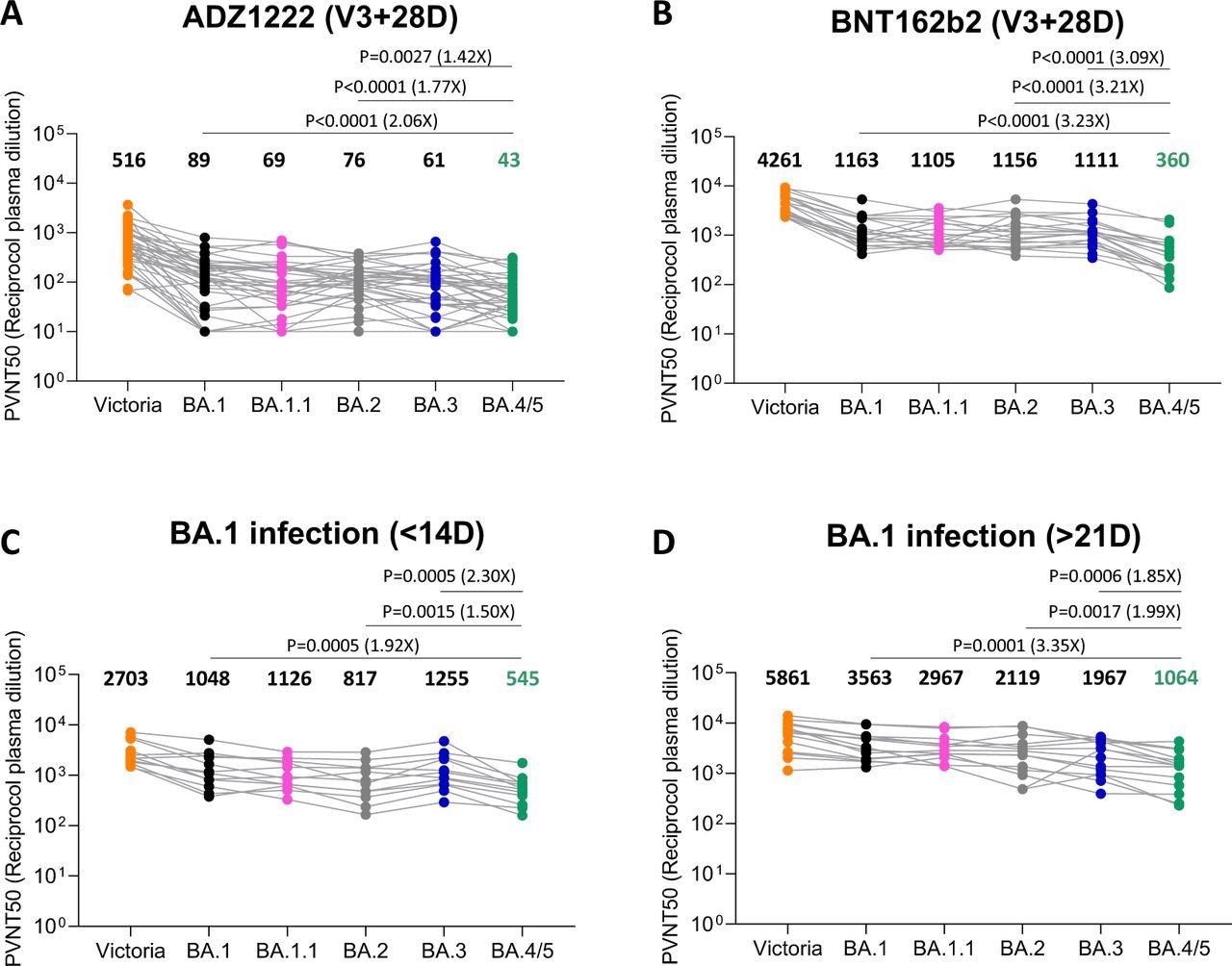
Pseudoviral neutralization assays of BA.4/5 by vaccine and BA.1 immune serum. IC50 values for the indicated viruses utilizing serum obtained from vaccinees 28 days following their third dose of vaccine (A) AstraZeneca AZD AZD1222 (n=41), (B) 4 weeks after the third dose of Pfizer BNT162b2 (n=20). Serum from volunteers struggling breakthrough BA.1 an infection volunteer taken (C) early ≤14 (n=12) days from symptom onset (median 13 days) (D) late ≥ 21 days from symptom onset (median 38 days) n=16. Comparability is made with neutralization titres to Victoria an early pandemic pressure, BA.1, BA.1..1, BA.2 and BA.3. Geometric imply titres are proven above every column. The Wilcoxon matched-pairs signed rank take a look at was used for the evaluation and two-tailed P values have been calculated.
In response to the AZD1222 vaccine, neutralization titers for BA.4/5 have been lowered 2.1-fold in comparison with BA.1 and 1.8-fold in comparison with BA.2. Likewise, the BNT162b2 vaccination lowered neutralization titers by 3.2-fold in comparison with each BA.1 and BA.2. This knowledge reveals that whereas vaccine efficacy in stopping an infection was low, notably at a later time level as antibody titers wane, it protected development in direction of extreme illness.
Nutalai et al. additionally generated a panel of 28 potent human monoclonal antibodies from sera of Omicron instances. Of those, 10 mAbs neutralized BA.4/5 utterly, whereas 4 different mAbs, Omi-09, 12, 29, and 35, confirmed a better than five-fold discount within the neutralization titer for BA.4/5 in comparison with BA.2. All of the mAbs interacted with the RBD, besides Omi-41, which sure the N-terminal area (NTD). Due to this fact, it neutralized BA.1, BA.1.1, and BA.3 particularly however not BA.2 or BA.4/5.
SPR evaluation confirmed a 1000-fold lower within the sensitivity of Omi-12 towards BA.4/5, an antibody delicate to the F486V mutation. Likewise, SPR confirmed a 77-fold enhanced neutralization of BA.4/5 in comparison with BA.2 for Omi-32, an L452R delicate antibody. The neutralizing exercise of AZD1061 was comparable towards BA.4/5 and BA.2, whereas the REG10987, which partially neutralized BA.2, was additional weakened towards BA.4/5.
The Omicron sub-lineages have been clustered collectively on the antigenic map however remained removed from earlier VOCs. Throughout the Omicron cluster, BA.4/5 was probably the most distant from the sooner sub-lineages, together with BA.1 and BA.2.
Conclusions
The research knowledge demonstrated that Omicron sub-lineages BA.4 and BA.5, in comparison with BA.1 and BA.2, escaped neutralization from sera of triple vaccinated people to a better extent. Thus, a brand new or repeat wave of Omicron an infection, pushed by BA.4/5, is probably going sooner or later. Furthermore, the neutralizing exercise of many mAbs was both knocked out or drastically impaired towards BA.4/5 in comparison with BA.2.
General, the research highlighted that there’s loads of antigenic area for SARS-CoV-2 to discover, and so it might evolve alongside the Omicron lineage or to a brand new variant, e.g., Delta to Omicron. SARS-CoV-2 is unlikely to be stopped by COVID-19 vaccination, however next-generation vaccines or vaccines together with pure immunity will make individuals proof against this extreme illness.
*Vital discover
bioRxiv publishes preliminary scientific experiences that aren’t peer-reviewed and, due to this fact, shouldn’t be thought to be conclusive, information scientific observe/health-related conduct, or handled as established data.
Journal reference:
- Additional antibody escape by Omicron BA.4 and BA.5 from vaccine and BA.1 serum, Aekkachai Tuekprakhon, Jiandong Huo, Rungtiwa Nutalai, Aiste Dijokaite-Guraliuc, Daming Zhou, Helen M. Ginn, Muneeswaran Sekvaraj, Chang Liu, Alexander J. Mentzer, Piyada Supasa, Helen M. E. Duyvesteyn, Raksha Das, Donal Skelly, Thomas G Ritter, Ali Amini, Sagida Bibi, Sandra Adele, Sile Ann Johnson, Bede Constantinides, Hermione Webster, Nigel Temperton, Paul Klenerman, Eleanor Barnes, Susanna J. Dunachie, Derrick Criminal, Andrew J. Pollard, Teresa Lambe, Philip Goulder, OPTIC consortium, ISARIC4C consortium, Elizabeth E Fry, Juthathip Mongkolsapaya, Jingshan Ren, David I. Stuart, Gavin R. Screaton, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.05.21.492554, https://www.biorxiv.org/content material/10.1101/2022.05.21.492554v1
[ad_2]



