
[ad_1]
In a current research posted to the medRxiv* pre-print server, researchers in the US developed a pandemic response commons (PRC) known as the Chicagoland coronavirus illness 2019 (COVID-19) commons (CCC). The CCC served Chicago, the state of Illinois, and surrounding areas in the US (US).
 Research: The Pandemic Response Commons. Picture Credit score: Orpheus FX / Shutterstock
Research: The Pandemic Response Commons. Picture Credit score: Orpheus FX / Shutterstock
Background
The US Facilities for Illness Management and Prevention (CDC) monitoring challenge pointed at a number of regional variations within the COVID-19 incidence, fatalities, and well being disparities. Due to this fact, it turned essential to curate, combine, and analyze COVID-19 information on the regional ranges and mixture the outcomes to tell national-level insurance policies.
An information commons, corresponding to PRC, curate, combine, and harmonize information for a selected group, e.g., researchers learning an epidemic or pandemic, public well being employees, and policymakers. Sometimes they require a number of authorized and information agreements.
Nevertheless, a regional occasion of a PRC developed within the present research was designed to be a part of a broader information ecosystem, function at a low degree, and enhance exercise as required by the pandemic. Most significantly, it comprised a number of regional commons to help the pandemic response via native, regional, and federated information sharing and evaluation.
Concerning the research
Within the current research, researchers used the open-source Gen3 information platform to develop PRC, and a proper consortium of Chicagoland space organizations operated it. Gen3, primarily based upon consortium, information, and platform agreements, was developed by the non-profit Open Commons Consortium.
The Open Commons Consortium has three fundamental features, as follows:
i) it helps institution of a consortium to construct and function a knowledge commons,
ii) ensures information is contributed to an information commons, and
iii) facilitates its members to work in teams, analyze information, and develop software program purposes and companies to reinforce the performance of the commons.
The CCC curated and harmonized a number of datasets, together with scientific information of ~90,000 sufferers, statistical information abstract of COVID-19 circumstances, and sequencing information of over 5,300 extreme acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant genomes.
Research findings
The CCC had eight members and 5 working teams. Its eight members, viz., Rush College Medical Middle, College of Chicago, Southern Illinois College, the College of Illinois at Chicago, St. Anthony Hospital, Sinai Chicago, NorthShore College HealthSystem, and CommunityHealth, had contributed scientific information from over 90,000 topics with COVID-19.
The scientific information working group developed a typical information mannequin for every member to contribute information within the required format. The epidemiological modeling working group used the CCC-obtained aggregated counts for COVID-19 circumstances, deaths, and choose comorbidities to know well being disparities and construct predictive fashions. They developed hierarchical Bayesian fashions that predicted county-wise future COVID circumstances and fatality counts for Illinois. Likewise, the working group developed regression fashions to know temporal, age-related race/ethnicity variations in case/fatality ratios. The variant surveillance working group collected and contributed over 5,300 SARS-CoV-2 genome sequences to nationwide and worldwide genomic databases.
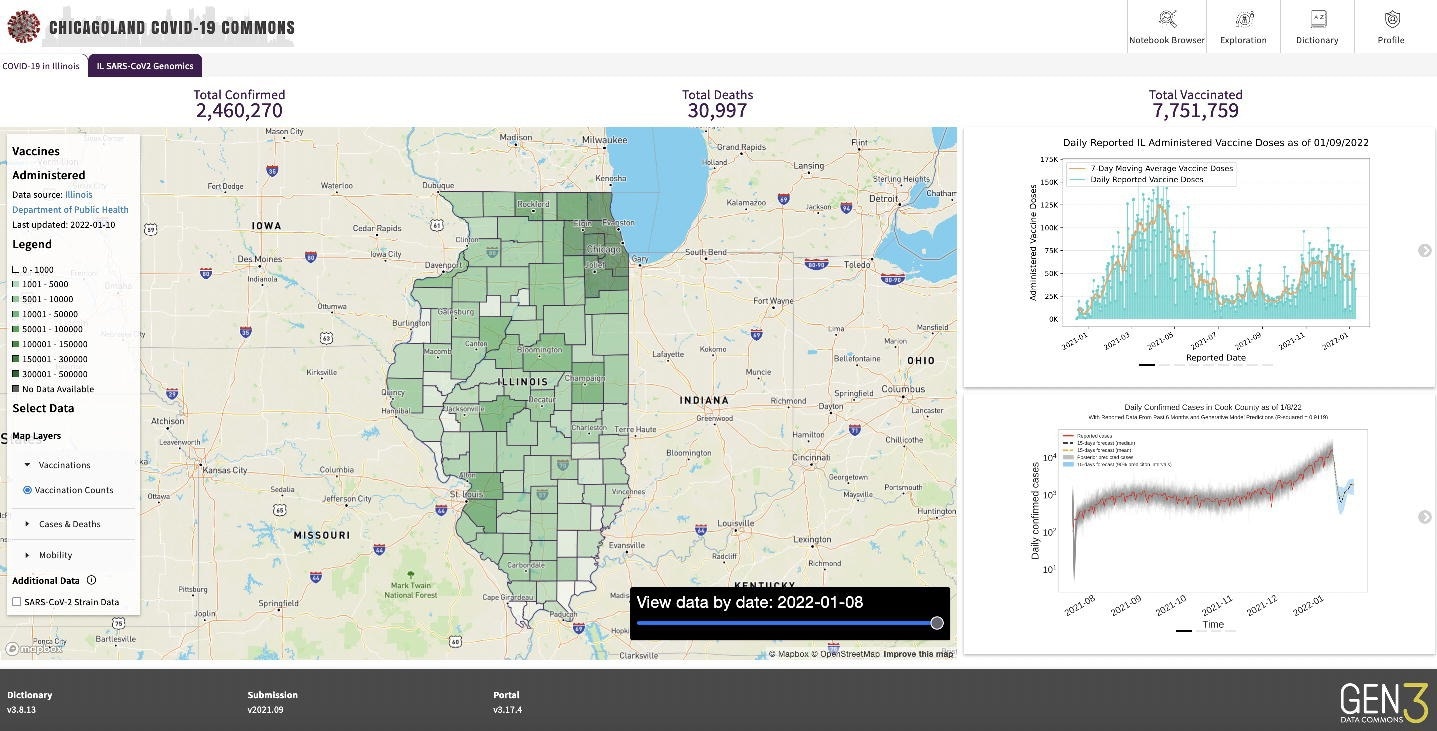 Screenshot of PRC
Screenshot of PRC
Gen3 software program routinely generates software programming interface (APIs) for information and metadata entry, information submission, authorization, and authentication, all of which make each managed and public entry findable, accessible, interoperable, and reusable (FAIR). As an illustration, PRC hosts a publicly accessible PRC Jupyter Pocket book Browser that helps entry COVID-19 case incidence, fatality, scientific, mobility, and imaging information.
Three taking part establishments contributed patient-level COVID-19 information beginning March 1, 2020. The PRC analyzed submitted information and recognized information high quality points. The standard evaluation included creating plots to match affected person counts by demography, signs, hospitalization occasions, and pre-existing comorbidities. Additional, the PRC used statistical abstract stories (SSR) county-level information to develop epidemiological fashions, which offered info for map overlays which might be simply accessible to the general public.
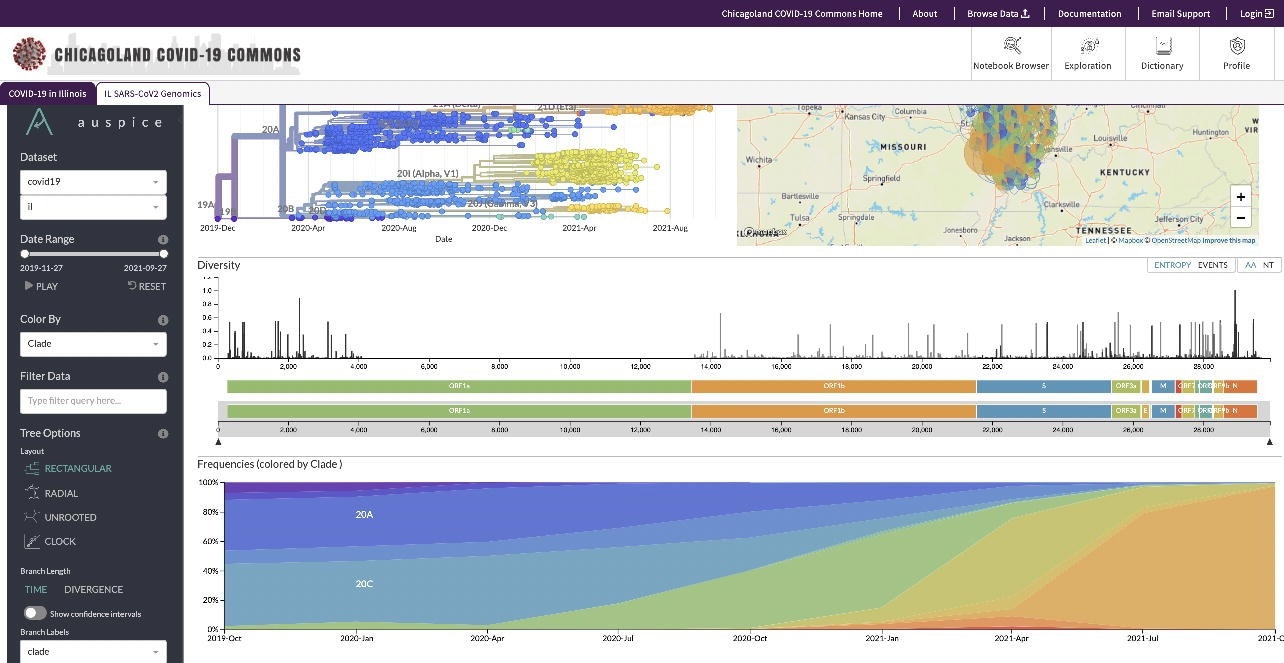
Screenshot of viral variants and their geographic distribution
The PRC additionally labored on a challenge with Southern Illinois College (SIU) to investigate the genomic sequence of SARS-CoV-2 and higher perceive the unfold of COVID-19 throughout Illinois. The challenge had sequenced over 5,300 SARS-CoV-2 genomes spanning 16 viral clades and greater than 150 variants to trace SARS-CoV-2 evolution in Illinois and establish the looks of particular SARS-CoV-2 variants of concern (VOCs).
Conclusions
The CCC contained scientific information from over 90,000 COVID-19, SSRs for the evaluation of COVID-19 well being disparities, over 5,300 SARS-CoV-2 genome sequencing information, and COVID-19-related public information. General, the CCC information was wealthy, available to a broader group, and enhanced the nationwide view of COVID-19-related points to speed up analysis on COVID-19 and Lengthy COVID. In abstract, the research highlighted the importance of a regional COVID-19 commons in complementing the continuing efforts to collect COVID-19 information on the nationwide degree to assist help scientific analysis and coverage improvement.
*Necessary discover
medRxiv publishes preliminary scientific stories that aren’t peer-reviewed and, subsequently, shouldn’t be considered conclusive, information scientific follow/health-related habits, or handled as established info.
Journal reference:
- The Pandemic Response Commons, Matthew Trunnell, Casey Frankenberger, Bala Hota, Troy Hughes, Plamen Martinov, Urmila Ravichandran, Nirav S Shah, Robert L Grossman, medRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.20.22276542, https://www.medrxiv.org/content material/10.1101/2022.06.20.22276542v1
[ad_2]



